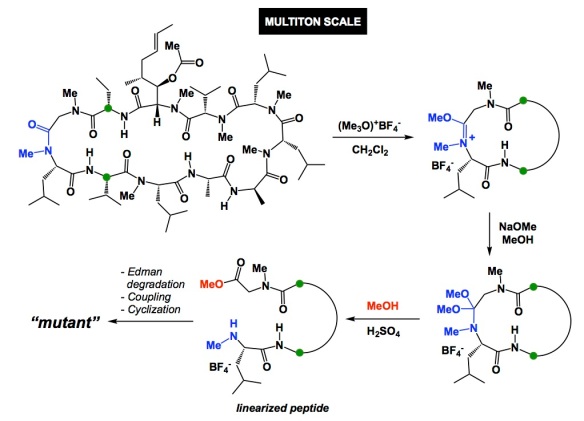
I was visiting Novartis in Basel, Switzerland, over the past 3 days. It is an amazing site, with astounding architecture. They even have a Frank Gehry building, a dinosaur skeleton, and a Japanese restaurant on campus. I went out for dinner with Dr. Fabrice Gallou last night and learned about the “business of cyclosporine A” at Novartis. The graphic below showcases the degree of sophistication achievable with complex molecules. The overall goal of the methylation reaction is to site-selectively cleave cyclosporine, run Edman degradation in order to remove the N-terminal amino acid, couple a new one, and recyclize. This represents a clever way of site-selectively mutate amino acids in complex macrocycles. The procedure discussed in the OPRD article was performed on a 186kg scale but things get even more impressive because the reaction is performed on a multi-ton scale at Novartis. As far as the site of methylation: it corresponds to the most nucleophilic amide. I wonder if more macrocycles do not possess an Achilles heel of this kind, which would allow site-selective chemistry to be applicable in other settings. I doubt it and, even if sites like this were to exist, they would likely be located in turn regions. Of particular interest to me was the description of effort to develop solvent/antisolvent combination in order to get consistent crystallinity of the linearized peptide product. 2-Methyltetrahydrofuran was the solvent and tert-butylmethyl ether was the antisolvent of choice. Go figure. I don’t think we use these two with linear peptides, but what do I know? Just imagine doing this on a multiton scale! Here is a quiz: why is cyclosporine’s OH acetylated? The authors discuss this, but try to guess the reason why having an unprotected OH is not a good idea.

Correction needed: in the linearized peptide, the methylene group in the sarcosine residue has gone AWOL.
Good call!